UDC 577.2:631:581.115:542.1 doi: 10.21498/2518-1017.19.4.2023.292911
Plant Breeding and Genetics Institute – National Center of Seed and Cultivar Investigation, 3 Ovidiopolska doroha St., Odesa, 65036, Ukraine, *e-mail: faygen@ukr.net
Purpose. Identification and evaluation of the frequencies of dominant and recessive alleles of the Ppd-A1 gene in winter and spring durum wheat varieties of different geographical origins. Methods. DNA isolation, allele-specific PCR, electrophoresis in agarose and polyacrylamide gels and statistical analysis were used in the research. Results. Using diagnostic molecular markers, the genotypes of 81 spring and winter durum wheat varieties from different geographical origins were identified by alleles of the Ppd-A1 gene, which determines differences in photoperiodic sensitivity. Four alleles were found in spring varieties and three in winter varieties (the dominant allele Ppd-A1a.2 was absent). The recessive allele Ppd-A1_del303 was not found in any of the examined varieties. Conclusions. No significant differences were found between winter and spring genotypes in the frequency of one or the other allele. In winter and spring varieties, the recessive allele Ppd-A1_del2ex7 is the most frequent (68.5 and 47.9%, respectively). The recessive allele Ppd-A1b is significantly lower in winter varieties and almost identical in spring varieties. The frequencies of the dominant alleles Ppd-A1a.2 and Ppd-A1a.3 are lower than the two above and generally very low. The Ppd-A1a.2 allele was detected only in the Georgian variety ‘Merliuri’ (spring type); Ppd-A1a.3 – in the Ukrainian varieties ‘Luhanska 7’, ‘Metyska’ (spring) and ‘Koralovyi’ (winter). The possibility of using varieties carrying the dominant alleles Ppd-A1a.2 and Ppd-A1a.3 as donors in hard winter wheat breeding programmes is currently being discussed, in order to increase their adaptive potential in conditions of drought and high temperatures and to increase grain yield. The use of marker analysis will ensure the selection of breeding material with the optimal combination of alleles of the Ppd-A1a gene.
Keywords: Triticum durum; type of development; photoperiod; Ppd-1 genes; genotype.
Viktor Fait
https://orcid.org/000000019994341X
Irina Balashova
https://orcid.org/0000000178551134
Introduction
Due to the significant variability of a trait such as heading time depending on seasonal conditions, wheat is able to minimise the impact of stress factors such as frost, heat, drought and others [1]. Optimising the heading time of plants by selecting appropriate alleles controlling this trait can improve adaptation and increase yield potential, and is a useful tool for selecting varieties suitable for cultivation in different conditions and geographical regions [2].
Photoperiodism is one of the most important natural mechanisms determining the length of the period before heading. The analysis of 79 modern varieties of winter durum wheat, originating from the Plant Breeding and Genetics Institute – National Center of Seed and Cultivar Investigation (PBGI – NCSCI), and foreign varieties adapted to the conditions of southern Ukraine, revealed only one variety with weak photoperiodic sensitivity, and most of the varieties studied had average or strong photoperiodic sensitivity [3]. Royo et al. [4] state that early flowering genotypes with the lowest photoperiodic sensitivity were the most productive when grown during several years of research in Spain, northern and southern Mexico. On the other hand, late flowering due to high photoperiodic sensitivity did not confer any advantage in grain yield. In durum wheat, photoperiod sensitivity is determined by the Ppd-A1 and Ppd-B1 genes located on chromosomes 2AS and 2BS, respectively [5]. Reduced photoperiod sensitivity in durum wheat results from promoter mutations in one of the two Ppd-1 genes. It is thought that the main differences in photoperiod response in T. durum are related to variability in the Ppd-A1 gene itself. Wilhelm et al. [6] identified two large deletions of 1027 and 1127 bp in the promoter of the tetraploid wheat varieties T. durum GS-100 and GS-105, which are designated as alleles Ppd-A1a.2 and Ppd-A1a.3, respectively. Damage to the promoter structure results in altered expression parameters, which persist throughout the day with the highest levels observed during the dark phase. A comparison of the nucleotide sequences of photoperiod-insensitive Ppd-A1a alleles, found in tetraploid wheat varieties T. durum GS-100 and GS-105, revealed that they arose independently of each other [7]. The high frequency of deletions of 1027 and 1127 bp in modern cultivated varieties, and their absence in wild tetraploid wheat, led to the conclusion that they arose during the domestication of wheat [8]. The wild-type Ppd-A1b allele, which is only expressed from dawn to the beginning of the dark phase, causes sensitivity to the shortening of the day length [6]. At the same time, it was found that the Ppd-A1 gene has mutations not only in the promoter, but also in the introns and exons. Such mutations can alter the genetic code, leading to the formation of Ppd proteins that partially or completely lose their function as inducers of the FT (Vrn-3) flowering locus. In particular, non-functional proteins are encoded by recessive alleles resulting from a
303-bp deletion, which covers the region of intron 5 and exons 5 and 6, as well as a 2-bp deletion in exon 7 of the Ppd-A1 gene. We have designated these alleles as Ppd-A1_del303 and Ppd-A1_del2ex7, respectively. In general, a significant number of mutations have been identified in the Ppd-A1 gene, grouped into more than 60 haplotypes [9].
The frequency of the insensitive allele Ppd-A1a.3 (variety GS105) was higher (34%) than alleles Ppd-A1a.2 (20%; variety GS100) among varieties of spring durum wheat [10]. In Argentine durum wheat varieties, the Ppd-A1a.3 allele (GS105 variety) had a frequency of only 25%, while the Ppd-A1b allele was present in 75% of genotypes [11]. Royo et al. [12] found the Ppd-A1_del303 allele in 20% of local varieties, but not in any of the modern ones. Only 1.3% of landraces possessed the photoperiod-insensitive allele Ppd-A1a.3 (variety GS105), and the allele Ppd-A1a.2 (GS100) was entirely absent. The group of modern varieties contained both sensitive and insensitive alleles.
The genotypes of carriers of individual alleles of the Ppd-A1 gene can be arranged in the following order based on the duration of the period before heading: Ppd-A1_del303, Ppd-A1b, Ppd-A1a.3 (GS105), and Ppd-A1a.2 (GS100) [12]. Furthermore, the Ppd-A1a.3 (GS105) allele was associated with an increase in harvest index, a decrease in plant height, and a decrease of protein content in grain. An increase in the number of grains per spike is associated with Ppd-A1a.3 (GS105), but also with smaller spikelets per spike [11]. Additionally, the Ppd-A1a.2 allele (GS100) contributes to a slight increase in the weight of one thousand grains and yield [12].
Currently, there is limited information on the distribution of dominant and recessive alleles of the Ppd-A1 locus in durum wheat varieties, as well as other cultivated types of wheat in Ukraine. Additionally, the influence of these alleles on the rates of development before heading has not been assessed to date.
The purpose of the research. Identification and evaluation of the frequencies of dominant and recessive alleles of the Ppd-A1 gene in winter and spring durum wheat varieties of different geographical origins.
Materials and methods
The starting material for this study consisted of ancient and modern varieties of durum wheat (Triticum durum Desf.). A total of 35 winter and 46 spring development types of different origins were used. Among the winter varieties, there were 33 varieties of Ukrainian breeding, including almost all of PBGI – NCSCI and the Plant Production Institute named after V. Ya. Yuriev, as well as single varieties from Romania (‘Pandur’) and Austria (‘Lupidur’). The spring sample included 23 breeding varieties from various Scientific Research Institutions in Ukraine, along with four varieties from Russia (‘Beloturka’, ‘Voronezhskaya 7’, ‘Donskaya Elegia’, ‘Novodonskaya’), three from Portugal (‘Marzaga’, ‘Presto De Tavira’, ‘Trems’), three Italy (‘Gumillo’, ‘Lumillo’, ‘Maliani 2’), two varieties from Georgia (‘Merliuri’ and ‘Tbilisuri 9’),
two varieties from France (‘Brindur’ and ‘Megadur’), several varieties from Azerbaijan (‘Shirvan 5’), Algeria (‘Oued Zenati 368’), Yeomen (‘Mestna’), Kazakhstan (‘Ema’), Mexico (‘Oviachic 65’), USA (‘Wells’), Tajikistan (‘Saodat’) and Hungary (‘Gk Basa’).
To determine the recessive and dominant alleles of the Ppd-A1 gene in T. durum, a multiplex polymerase chain reaction (PCR) was used (Table 1). The presence of an amplification fragment of 452 bp indicates the absence of any deletion in the promoter of the Ppd-A1 gene and, accordingly, the presence of a recessive allele. Fragments of 380 bp and 290 bp determine the dominant alleles Ppd-A1a.2 and Ppd-A1a.3, respectively [6]. Detection of a recessive allele with a 303 bp deletion in exons 5 and 6, designated as Ppd-A1_del303, and the allele Ppd-A1_del2ex7, which has a 2 bp deletion in exon 7, was carried out using the PCR markers recommended by S. Takenaka and T. Kawahara [14]. The sequence of primers is shown in Table 1. The presence of the Ppd-A1_del303 allele is detected by a fragment of 220 bp, with the Capelle-Desprez variety being the reference sample that has such a mutation. The presence of the Ppd-A1_del2ex7 allele in the genotype is indicated by the presence of a 170 bp amplification fragment.
The reaction buffer for multiplex PCR amplification consisted of 50 mM KCl; 20 mM Tris-HCl, pH 8.4; 2.0 mM MgCl2; 0.01% Tween-20, 0.15 mM of each dNTP; 5 pM of each primer; 20 ng of DNA, and 1 unit of Taq polymerase. The reaction mixture volume was 20 μl. Amplification conditions: denaturation – 94 °C – 2 min, then – 20 s; annealing – 60 °C – 30 s; synthesis – 72 °C – 50 s. 35 cycles; the last elongation – 72 °C – 3 min.
Agarose and polyacrylamide (PAA) gels were used to test amplification products. The PCR products were separated on agarose gels, stained with ethidium bromide, and photographed under UV light. The PAA gel consisted of 10% acrylamide and 1 × TVE buffer, which is composed of 50 mM Tris-H3BO3 and 2 mM Na3EDTA at a pH of 8.0. Electrophoresis was carried out at a voltage of 500 V for 120 minutes at 60 °C. The gel was prepared by mixing
10 ml of 30% polyacrylamide solution, 3 ml of 10× TVE buffer, 25 μl of TMED, and 50 μl of 10% PSA. To control the movement of DNA fragments in the gel, each DNA sample was mixed with 4 μl of a 0.2% (w/v) solution of bromophenol blue and 0.2% (w/v) xylenecyanol. Ledder 1000 or pUC19/Msp1 was used to control the molecular weight of the amplified fragments.
Results and discussion
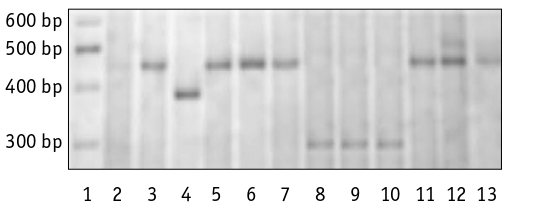
The genotypes of 35 durum winter wheat varieties and 46 spring wheat varieties were identified based on the alleles of the Ppd-A1 gene. Polymorphism of amplification fragments was observed among both winter and spring varieties (Fig. 1) The winter variety ‘Koralovyi’ and the spring varieties ‘Luhanska 7’ and ‘Metyska’ produced a PCR product with a size of 290 bp, while the spring variety ‘Merliuri’ produced a 380 bp product. The first three genotypes carry the dominant allele Ppd-A1a.3, while the ‘Merliuri’ variety carries the Ppd-A1a.2 allele (Table 2). There is no significant difference in the proportion of varieties carrying either the Ppd-A1a.2 or Ppd-A1a.3 allele between winter and spring varieties. The difference in the proportion of varieties with different types of development was only 2.2 ± 3.40% in the first case and 1.4 ± 5.18% in the second. The electrophoreogram showed the presence of a 452 bp amplification fragment in all 77 other genotypes, regardless of their developmental type. This suggests that one of the recessive alleles of the Ppd-A1 gene is present in their genotypes.

Fig. 1. Electropherogram of multiplex PCR products detecting Ppd-A1b, Ppd-A1a.2, Ppd-A1.3 alleles in T. durum varieties:
1 – Ledder 1000; 2 – ‘Kolektyvna 2’, 3 – ‘Kharkivska 46’,
4 – ‘Merliuri’, 5 – ‘Spadshchyna’, 6 – ‘Chado’, 7 – ‘Narodna’,
8 – ‘Metyska’, 9 – ‘Luhanska 7’, 10 – ‘Koralovyi’,
11 – ‘Akveduk’, 12 – ‘Blyskuchyi’, 13 – ‘Linkor’
The PCR test was unable to detect the presence of the mutant recessive allele, Ppd-A1_del303, which results in a non-functional protein due to a violation of the structure of intron 5 and exons 5 and 6 of the Ppd-A1 gene, in any of the varieties in the general sample. However, Royo et al. [12] reported the presence of this allele in local varieties of spring durum wheat in Mediterranean countries, but not in any modern commercial variety. The Ppd-A1_del303 allele is prevalent in modern soft winter wheat from north-western Europe, particularly in Sweden, as well as in old local varieties [15]. This suggests that the allele may have been selected for these growing conditions. However, it is possible that this prevalence is due to the founder effect or historical selection of linked genes with favourable alleles for other traits. Further study is required to determine the effects of this allele in different conditions.The Ppd-A1_del2ex7 allele is prevalent among the studied varieties
(Fig. 2). It was identified in 57% (46 samples), with a share of 47.8% in spring varieties and 68.5% in winter varieties. However, the differences between these two groups of varieties were not significant (t = 1.92 at t0.05 = 2.01).
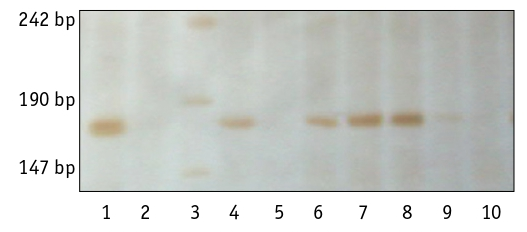
Fig. 2. Marking of the allele Ppd-A1_del2ех7 of durum wheat varieties:
1, 4, 6, 7, 8 – the presence of a mutant allele in the varieties ‘Parus’, ‘Aisberh’, ‘Kolektyvna 2’, ‘Kharkivska 46’, ‘Biloturka’; 2, 5, 9, 10 – the absence of a mutant allele
in the varieties ‘Metyska’, ‘Merliuri’, ‘Yaskravyi’, ‘Chado’;
3 –molecular weight marker pUC19/Msp1
Of the remaining studied varieties, 31 or 38.3% possess the “classic” recessive Ppd-A1b allele in their genotype. The number of these genotypes in winter varieties is more than two times less than in spring varieties. However, the differences in the frequency of Ppd-A1b allele distribution between groups of varieties of different developmental types are not significant (t = 1.61 at t0.05 = 2.04). Based on marker analysis of 81 durum wheat varieties, it was found that 77 of them (95.1%) carry two different recessive alleles, while only four samples (4.9%) carry two different dominant alleles of the Ppd-A1 gene. The proportions of the two groups of genotypes of carriers of different dominant alleles Ppd-A1a.2 or Ppd-A1a.3 did not differ from each other in the general sample (t = 1.04 with t0.05 = 2.78) or in the samples of winter (t = 0.75 with t0.05 = 17.71) and spring (t = 0.57 with t0.05 = 3.18) varieties. Meanwhile, the proportion of genotypes carrying the recessive allele Ppd-A1_del2ex7 significantly exceeds that of the Ppd-A1b genotype by almost 40% (t = 3.64 at t0.05 = 2.04) in the winter varieties. However, in the spring varieties, this advantage is only 2.1% and is not significant (t = 0.19 at t0.05 = 2.02). It is important to note that in durum wheat, which has a photoperiodic response controlled by only two Ppd-1 genes, nearly half of the varieties examined had the Ppd-A1 gene with a mutation that results in the loss of Ppd protein functionality. In the same time, the frequencies of the recessive alleles Ppd-A1_del2ex7 and Ppd-A1b significantly exceeded those of the dominant alleles Ppd-A1a.2 or Ppd-A1a.3 by 41.4–45.6% in spring varieties (t = 5.23–7.50 with t0.05 = 2.07) and by 25.7–68.5% in winter varieties (t = 2.24–7.86 with t0.05 = 2.06–2.23).
Out of the 57 Ukrainian durum wheat varieties, 33 are winter and 24 are spring. None of these varieties were found to carry the dominant Ppd-A1a.2 allele. The percentage of carriers of the dominant Ppd-A1a.3 allele is very low, at around 8% for spring and 3% for winter. Therefore, over 90% of genotypes for both spring and winter varieties of durum wheat carry recessive alleles. The presented findings contradict the previous results on the ratio of sensitive and insensitive photoperiod genotypes in samples of winter and spring varieties of bread wheat of Ukrainian breeding [16]. Specifically, the proportion of varieties carrying recessive alleles of Ppd-1 genes is several times higher in spring varieties of bread wheat than in winter varieties, and vice versa in the sample of winter varieties – dominant alleles. The absence of dominant alleles in spring varieties of durum wheat and bread wheat may be due to the cultivation area and sowing time. In Ukraine, spring wheat is usually cultivated in the northern regions with sowing taking place between late March and early April. The primary stages of spring wheat development occur during late spring and summer, with longer natural daylight hours (15–16 hours), which largely eliminates differences in the effects of photoperiod genes. Under these conditions, genotypes with dominant alleles of Ppd-1 genes no longer have a significant advantage in terms of grain yield.
The growth and development of winter varieties, both bread and durum, falls on the autumn-winter and early spring periods, when the length of the natural day in all regions of our country is insufficient (9–11 hours) for the development of wheat. As a result, the transition to heading in photoperiod-sensitive varieties occurs quite late, which increases the probability of winter durum wheat plants falling under the influence of heat and drought in the second part of the growing season, which is significantly intensified during the period of ripening and filling of grain throughout the territory of Ukraine, especially in the south. It has been proven that insensitivity to the photoperiod is useful for bread winter wheat in southern regions with high summer temperatures [17–19], including the south of Ukraine [20, 21]. At the same time, among Ukrainian winter durum wheat varieties, only ‘Koralovyi’ variety is a carrier of the dominant Ppd-A1a.3 allele.
The absence of dominant alleles of the Ppd-A1a gene in winter durum wheat varieties may be attributed to their origin. Winter durum wheat is typically created through interspecific hybridization of spring durum and winter bread wheat, followed by intraspecific hybridisation [22]. The distribution of dominant alleles of the Ppd-A1 gene is significantly limited among spring varieties of durum wheat, and its share in different collections of winter varieties of bread wheat varies from 0 to 15% [23–25]. However, the introgression of dominant alleles of the Ppd-A1 gene into newly created varieties can reduce the negative impact of climate change on durum wheat grainfilling [26].
Conclusions
Genotypes of 81 varieties of spring and winter durum wheat were identified by alleles of the Ppd-A1 gene. Four alleles were found in spring varieties and three in winter varieties. The latter lack the Ppd-A1a.2 allele. No recessive allele Ppd-A1_del303 was detected in any variety, regardless of the type of development.
No significant differences were found between winter and spring genotypes in the distribution frequency of each of the four alleles. In winter and spring varieties, the most frequent recessive allele was Ppd-A1_del2ex7 (68.5 and 47.9%, respectively), whose frequency was significantly higher than the other recessive allele Ppd-A1b in winter varieties and almost equal to that in spring varieties.
The frequencies of the dominant alleles Ppd-A1a.2 and Ppd-A1a.3 are quite low and significantly lower than those of the alleles Ppd-A1_del2ex7 and Ppd-A1b. The Ppd-A1a.2 allele was detected only in the Georgian spring variety ‘Merliuri’ and the Ppd-A1a.3 allele in the Ukrainian spring varieties ‘Luhanska 7’ and ‘Metyska’ and the winter variety ‘Koralovyi’.
The use of varieties carrying the dominant alleles Ppd-A1a.2 and Ppd-A1a.3 as donors in winter durum wheat breeding programmes will contribute to increasing the adaptive potential to heat and high temperature conditions and to increasing grain yield, and the use of marker analysis can ensure the selection of breeding material with an optimal combination of alleles of the Ppd-A1a gene.
References
Використана література
Table 1
Primer sequences and expected sizes of PCR products
for marking alleles of the Ppd-A1 gene
|
Gene |
Primer |
Primer sequence |
Amplifier size |
|
Ppd-A1b / Ppd-A1a.2 / Ppd-A1a.3 |
durum_AglF1 |
gtatgcgattcgcctgaagt |
452/380/290 bp [6] |
|
durum_AglF2 |
cgtcacccatgcactctgtt |
||
|
durum_AglR2 |
ctggctccaagaggaaacac |
||
|
Ppd-A1_del303 |
303 bp_del_F2 |
cttacatctgtgagaagtatctgcatc |
220 bp [14] |
|
303 bp_del_R3 |
cagatcagcagctcgaacaattac |
||
|
Ppd-A1_del2ex7 |
2 bp_del_F1 |
gccgccgtgaacaagttg |
170 bp [14] |
|
2 bp_del_R1 |
ggtaacgcacctgcaaaatgag |
Table 2
Genotypes of hard winter and spring wheat varieties from different geographical regions
and their allele frequencies at the Ppd-A1 locus
|
Genotype |
Type of development |
Variety |
n |
P ± Sp, % |
Together |
|
|
n |
p ± Sp, % |
|||||
|
Ppd-А1a.2 |
winter |
|
0 |
0.0 ± 2.63 |
1 |
1.2 ± 1.21 |
|
spring |
‘Merliuri’ |
1 |
2.2 ± 2.16 |
|||
|
Ppd-А1а.3 |
winter |
‘Koralovyi’ |
1 |
2.9 ± 2.84 |
3 |
3.7 ± 2.09 |
|
spring |
‘Luhanska 7’, ‘Metyska’ |
2 |
4.3 ± 2.99 |
|||
|
Ppd-A1b |
winter |
‘Akveduk’, ‘Arhonavt’, ‘Burshtyn’, ‘Hordeiforme 3’, ‘Kontynent’, ‘Kreiser’, ‘Lainer’, ‘Prestyzhnyi’, ‘Kharkivska 1’, ‘Yaskravyi’ |
10 |
28.6 ± 7.64 |
31 |
38.3 ± 5.40 |
|
spring |
‘Donskaya elegiya’, ‘Ema’, ‘Zhyzel’, ‘Izolda’, ‘Mestna’, ‘Kharkivska 3’, ‘Kharkivska 13’, ‘Kharkivska 15’, ‘Kharkivska 25’, ‘Kharkivska 51’, ‘Chado’, ‘Shyrvan 5’, ‘Shovkovysta’, ‘Brindur’, ‘Gk Basa’, ‘Gumillo’, ‘Marzaga’, ‘Megadur’, ‘Oviachic 65’, ‘Presto De Tavirа’, ‘Oued Zenati 368’ |
21 |
45.7 ± 7.34 |
|||
|
Ppd-A1_del2ex7 |
winter |
‘Aysberh’, ‘Alyy parus’, ‘Almaznyy’, ‘Areal’, ‘Afina’, ‘Blyskuchyi’, ‘Bosfor’, ‘Havan’, ‘Hardemaryn’, ‘Delfin’, ‘Zolote Runo’, ‘Lahuna’, ‘Linkor’, ‘Makar’, ‘Nadiinyi’, ‘Parus’, ‘Perlyna’, ‘Prybutkova’, ‘Prozoryi’, ‘Tur’, ‘Faktor Odeskyi’, ‘Shliakhetnyi’, ‘Lupidur’, ‘Pandur’ |
24 |
68.5 ± 7.85 |
46 |
56.8 ± 5.50 |
|
spring |
‘Arnautka’, ‘Beloturka’, ‘Voronezhskaya 7’, ‘Dunyasha’, ‘Kolektyvna 2’, ‘Kuchumivka’, ‘Lumillo’, ‘Narodna’, ‘Novodonskaya’, ‘Prykrasa’, ‘Saodat’, ‘Tbilisuri 9’, ‘Tera’, ‘Kharkivska 21’, ‘Kharkivska 33’, ‘Kharkivska 37’, ‘Kharkivska 39’, ‘Kharkivska 46’, ‘Chornokoloska’, ‘Maliani 2’, ‘Trems’, ‘Wells’ |
22 |
47.8 ± 7.36 |
|||
|
Total |
winter |
– |
35 |
100 |
81 |
100 |
|
spring |
– |
46 |
100 |
|||
УДК 577.2:631:581.115:542.1
Файт В. І.*, Балашова І. А. Різноманіття сортів ярої та озимої пшениці твердої (Triticum durum Desf.) за алелями гена PpdА1. Plant Varieties Studying and Protection. 2023. Т. 19, № 4. С. –. https://doi.org/10.21498/25181017.19.4.2023.292911
Селекційногенетичний інститут – Національний центр насіннєзнавства та сортовивчення, Овідіопольська дорога, 3, м. Одеса, Україна, 65036, *email: faygen@ukr.net
Мета. Ідентифікувати та оцінити частоти домінантних і рецесивних алелів гена Ppd-A1 в озимих та ярих сортів пшениці твердої різного географічного походження. Методи. Під час досліджень використовували методи виділення ДНК, алель-специфічної ПЛР, електрофорезу в агарозному й поліакриламідному гелях, статистичного аналізу. Результати. Завдяки застосуванню діагностичних молекулярних маркерів ідентифіковано генотипи 81 сорту твердої пшениці ярого та озимого типів розвитку різного географічного походження за алелями гена Ppd-A1, що визначає відмінності за фотоперіодичною чутливістю. В ярих сортів виявлено чотири алелі, в озимих –
три (відсутній домінантний алель Ppd-А1a.2). В жодного сорту незалежно від типу розвитку не ідентифіковано рецесивного алеля Ppd-A1_del303. Висновки. Істотних відмінностей між озимими та ярими генотипами за частотою того чи іншого алеля не встановлено. Серед озимих і ярих сортів найпоширенішим є рецесивний алель Ppd-A1_del2ex7 (68,5 і 47,9% відповідно). Значно йому поступається в озимих сортів і майже однаковий з ним за частотою розповсюдження в ярих рецесивний алель Ppd-A1b. Меншими за дві вищевказані й загалом дуже малими є частоти домінантних алелів Ppd-А1a.2 і Ppd-А1а.3. Алель Ppd-А1a.2 виявлено лише у грузинського сорту ‘Мерліурі’ (ярий тип розвитку); Ppd-А1а.3 – в українських сортів ‘Луганська 7’, ‘Метиска’ (ярі) та ‘Кораловий’ (озимий). Натепер обговорюють можливість використання сортів-носіїв домінантних алелів Ppd-А1a.2 і Ppd-А1а.3 як донорів у програмах селекції пшениці твердої озимої з метою підвищення її адаптивного потенціалу в умовах посухи та високих температур і для збільшення врожаю зерна. Застосування маркерного аналізу дасть змогу забезпечити добір селекційного матеріалу з оптимальною комбінацією алелів гена Ppd-А1a.
Ключові слова: Triticum durum; тип розвитку; фотоперіод; Ppd-1 гени; генотип.
Надійшла / Received 02.11.2023
Погоджено до друку / Accepted 20.11.2023